What’s the Buzz About CRISPRs?
Sometimes, new ideas come from the strangest of places; inspired by something that seems completely unrelated. This sort of out-of-the-blue brain blast is exactly what happened in 2012 when a study about bacteria’s adaptive immunity opened up the possibility for a new approach to gene therapy. While CRISPR technology was runner-up for the Breakthrough Technology of the Year in 2012, its presence in bacteria cells has been known since 1987. It was not until a series of studies between 2005 and 2007, however, that their role in the bacterial adaptive immunity was revealed.
Short for Clustered Regularly Interspaced Short Palindromic Repeats; a CRISPR is a small length of DNA with a repeating and reversing sequence of base pairs (the pieces that make up genetic code). Found in a specific section (or loci) in the bacterial genome, multiple CRISPRs are usually grouped together in a string. Each CRISPR is followed by a ‘spacer’ section of DNA.
When pathogens, such as a virus, attack bacteria they insert a section of DNA into their victim. Oblivious to any danger the victim reads and processes this foreign DNA. In so doing, the bacteria also retain a record within its spacers enabling it to recognize and fight off the same pathogen in the future. Each spacer therefore acts as a ‘bookmark’ for any pathogen attack the cell has encountered.
How do bacteria do this? Preceding a CRISPR sequence are cas genes which make enzymes that copy the foreign DNA sequence and insert it as small fragments into the bacterial genome as new spacers.
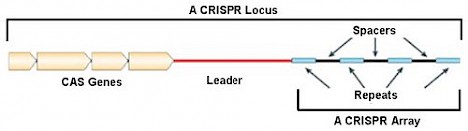
With this new spacer now available in the CRISPR loci, the next time a bacteria encounters the same attack it will recognize the foreign DNA and send out a secondary Cas protein to target, bind, and splice out the DNA thereby inactivating the invader, analogous to the way in which our bodies produce antibodies against repeated infections
It’s this ability to identify, isolate, and splice out a specific piece of DNA that has captured the imagination of scientists.
The CRISPR/Cas system can now be hijacked to accurately target genetic errors in DNA and remove them. Once removed repair systems kick in to fix the sequence. This can be done either of two ways: non-homologous end-joining which simply connects the two ends together or the more sophisticated homology-directed repair.
Applying this to Rett Syndrome we can use the following example. The most common mutation in MECP2 is the T158M where a cytosine to thymine
error at nucleotide base number 473 swaps a methionine amino acid for a threonine. One could envision using the CRISPR/Cas technology to introduce a cut at nucleotide base number 473, splice out the thymine and provide a cytosine base via a template. Cas enzymes and base repair templates would be delivered via gene therapy vectors.
So where does the technology currently stand to be able to achieve this? Inducing the break in DNA can now be achieved efficiently and effectively however the repair step using templates is not yet ready for prime time as efficiency rates (the number of cells that actually achieve repair) are still quite small. Before the technology can be considered for therapeutic applications it also needs to be shown that the CRISPR/Cas machinery only cuts the DNA at desired sites as additional “off target” breaks could be damaging.
The encouraging news is that progress with CRISPR/Cas technology is occurring at lightning speed with thousands of laboratories around the world working to improve the process. It is not difficult to imagine the immense possibilities for treating genetic disease. No wonder there is so much excitement with scientists themselves routinely referring to this new technology as revolutionary.


